CHAPTER 83 Table 83–1 classifies the major antimicrobial drugs according to susceptible organisms. The table shows three major groups: antibacterial drugs, antifungal drugs, and antiviral drugs. In addition, the table subdivides the antibacterial drugs into narrow-spectrum and broad-spectrum agents, and indicates the principal classes of bacteria against which they are active. TABLE 83–1 Classification of Antimicrobial Drugs by Susceptible Organisms ANTIBACTERIAL DRUGS Narrow Spectrum Gram-Positive Cocci and Gram-Positive Bacilli Penicillin G and V Penicillinase-resistant penicillins: methicillin, nafcillin Vancomycin Erythromycin Clindamycin Gram-Negative Aerobes Aminoglycosides: gentamicin, others Cephalosporins (first and second generations) Mycobacterium tuberculosis Isoniazid Rifampin Ethambutol Pyrazinamide Broad Spectrum Gram-Positive Cocci and Gram-Negative Bacilli Broad-spectrum penicillins: ampicillin, others Extended-spectrum penicillins: carbenicillin, others Cephalosporins (third generation) Tetracyclines: tetracycline, others Carbapenems: imipenem, others Trimethoprim Sulfonamides: sulfisoxazole, others Fluoroquinolones: ciprofloxacin, others ANTIVIRAL DRUGS Drugs for HIV Infection Reverse transcriptase inhibitors: zidovudine, others Protease inhibitors: ritonavir, others Fusion inhibitors: enfuvirtide Integrase inhibitors: raltegravir CCR5 antagonists: maraviroc Drugs for Influenza Adamantanes: amantadine, others Neuraminidase inhibitors: oseltamivir, others Other Antiviral Drugs Acyclovir Ribavirin Interferon alfa ANTIFUNGAL DRUGS Polyene antibiotics: amphotericin B, others Azoles: itraconazole, others Echinocandins: caspofungin, others The antimicrobial drugs fall into seven major groups based on mechanism of action. This classification is summarized in Table 83–2. Properties of the seven major classes are discussed briefly below. TABLE 83–2 Classification of Antimicrobial Drugs by Mechanism of Action • Drugs that inhibit bacterial cell wall synthesis or activate enzymes that disrupt the cell wall—These drugs (eg, penicillins, cephalosporins) weaken the cell wall and thereby promote bacterial lysis and death. • Drugs that increase cell membrane permeability—Drugs in this group (eg, amphotericin B) increase the permeability of cell membranes, causing leakage of intracellular material. • Drugs that cause lethal inhibition of bacterial protein synthesis—The aminoglycosides (eg, gentamicin) are the only drugs in this group. We do not know why inhibition of protein synthesis by these agents results in cell death. • Drugs that cause nonlethal inhibition of protein synthesis—Like the aminoglycosides, these drugs (eg, tetracyclines) inhibit bacterial protein synthesis. However, in contrast to the aminoglycosides, these agents only slow microbial growth; they do not kill bacteria at clinically achievable concentrations. • Drugs that inhibit bacterial synthesis of DNA and RNA or disrupt DNA function—These drugs inhibit synthesis of DNA or RNA by binding directly to nucleic acids or by interacting with enzymes required for nucleic acid synthesis. They may also bind with DNA and disrupt its function. Members of this group include rifampin, metronidazole, and the fluoroquinolones (eg, ciprofloxacin). • Antimetabolites—These drugs disrupt specific biochemical reactions. The result is either a decrease in the synthesis of essential cell constituents or synthesis of nonfunctional analogs of normal metabolites. Examples of antimetabolites include trimethoprim and the sulfonamides. • Drugs that suppress viral replication—Most of these drugs inhibit specific enzymes—DNA polymerase, reverse transcriptase, protease, integrase, or neuraminidase—required for viral replication and infectivity. Over time, an organism that had once been highly sensitive to an antibiotic may become less susceptible, or it may lose drug sensitivity entirely. In some cases, resistance develops to several drugs. Acquired resistance is of great concern in that it can render currently effective drugs useless, thereby creating a clinical crisis and a constant need for new antimicrobial agents. As a rule, antibiotic resistance is associated with extended hospitalization, significant morbidity, and excess mortality. Organisms for which drug resistance is now a serious problem include Enterococcus faecium, Staphylococcus aureus, Enterobacter species, Pseudomonas aeruginosa, Acinetobacter baumanii, Klebsiella species, and Clostridium difficile (Table 83–3). Two of these resistant bacteria—methicillin-resistant Staph. aureus and C. difficile—are discussed at length in Chapter 84 (Box 84–1) and Chapter 85 (Box 85–1), respectively. TABLE 83–3 Drugs for Some Highly Resistant Bacteria PBP5 = penicillin-binding protein 5. *Methicillin-resistant Staphylococcus aureus is discussed at length in Chapter 84 (see Box 84–1). †Clostridium difficile infection is discussed at length in Chapter 85 (see Box 85–1). New delhi metallo-beta-lactamase 1 (ndm-1) gene. Extensive drug resistance is conferred by the NDM-1 gene, which codes for a powerful form of beta-lactamase. As discussed in Chapters 84 and 85, beta-lactamases are enzymes that can inactivate drugs that have a beta-lactam ring. The form of beta-lactamase encoded by NDM-1 is both unusual and troubling in that it can inactivate essentially all beta-lactam antibiotics, a group that includes penicillins, cephalosporins, and carbapenems. Worse yet, the DNA segment that contains the NDM-1 gene also contains genes that code for additional resistance determinants, including drug efflux pumps, and enzymes that can inactivate other important antibiotics, including erythromycin, rifampicin, chloramphenicol, and fluoroquinolones. Furthermore, all of these genes are present on a plasmid, a piece of DNA that can be easily transferred from one bacterium to another (see below). Of note, bacteria that have the NDM-1 gene are resistant to nearly all antibiotics, except for tigecycline and colistin. Since its discovery in Klebsiella pneumoniae in 2008, NDM-1 has been found in other common enteric bacteria, including Escherichia coli, Enterobacter, Salmonella, Citrobacter freundii, Providencia rettgeri, and Morganella morganii. To date, only a few cases of NDM-1 infection have been reported in the United States and Canada.
Basic principles of antimicrobial therapy
Classification of antimicrobial drugs
Classification by susceptible organism

Classification by mechanism of action

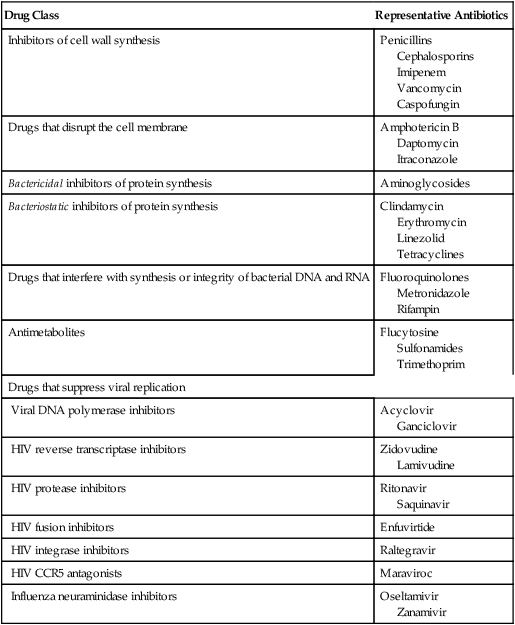
Drug Class
Representative Antibiotics
Inhibitors of cell wall synthesis
Penicillins
Cephalosporins
Imipenem
Vancomycin
Caspofungin
Drugs that disrupt the cell membrane
Amphotericin B
Daptomycin
Itraconazole
Bactericidal inhibitors of protein synthesis
Aminoglycosides
Bacteriostatic inhibitors of protein synthesis
Clindamycin
Erythromycin
Linezolid
Tetracyclines
Drugs that interfere with synthesis or integrity of bacterial DNA and RNA
Fluoroquinolones
Metronidazole
Rifampin
Antimetabolites
Flucytosine
Sulfonamides
Trimethoprim
Drugs that suppress viral replication
Viral DNA polymerase inhibitors
Acyclovir
Ganciclovir
HIV reverse transcriptase inhibitors
Zidovudine
Lamivudine
HIV protease inhibitors
Ritonavir
Saquinavir
HIV fusion inhibitors
Enfuvirtide
HIV integrase inhibitors
Raltegravir
HIV CCR5 antagonists
Maraviroc
Influenza neuraminidase inhibitors
Oseltamivir
Zanamivir
Acquired resistance to antimicrobial drugs

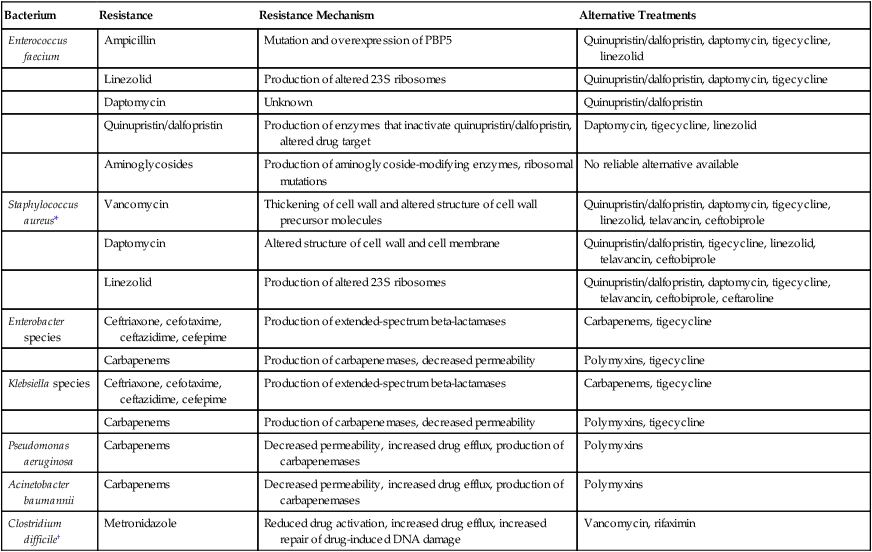
Bacterium
Resistance
Resistance Mechanism
Alternative Treatments
Enterococcus faecium
Ampicillin
Mutation and overexpression of PBP5
Quinupristin/dalfopristin, daptomycin, tigecycline, linezolid
Linezolid
Production of altered 23S ribosomes
Quinupristin/dalfopristin, daptomycin, tigecycline
Daptomycin
Unknown
Quinupristin/dalfopristin
Quinupristin/dalfopristin
Production of enzymes that inactivate quinupristin/dalfopristin, altered drug target
Daptomycin, tigecycline, linezolid
Aminoglycosides
Production of aminoglycoside-modifying enzymes, ribosomal mutations
No reliable alternative available
Staphylococcus aureus*
Vancomycin
Thickening of cell wall and altered structure of cell wall precursor molecules
Quinupristin/dalfopristin, daptomycin, tigecycline, linezolid, telavancin, ceftobiprole
Daptomycin
Altered structure of cell wall and cell membrane
Quinupristin/dalfopristin, tigecycline, linezolid, telavancin, ceftobiprole
Linezolid
Production of altered 23S ribosomes
Quinupristin/dalfopristin, daptomycin, tigecycline, telavancin, ceftobiprole, ceftaroline
Enterobacter species
Ceftriaxone, cefotaxime, ceftazidime, cefepime
Production of extended-spectrum beta-lactamases
Carbapenems, tigecycline
Carbapenems
Production of carbapenemases, decreased permeability
Polymyxins, tigecycline
Klebsiella species
Ceftriaxone, cefotaxime, ceftazidime, cefepime
Production of extended-spectrum beta-lactamases
Carbapenems, tigecycline
Carbapenems
Production of carbapenemases, decreased permeability
Polymyxins, tigecycline
Pseudomonas aeruginosa
Carbapenems
Decreased permeability, increased drug efflux, production of carbapenemases
Polymyxins
Acinetobacter baumannii
Carbapenems
Decreased permeability, increased drug efflux, production of carbapenemases
Polymyxins
Clostridium difficile†
Metronidazole
Reduced drug activation, increased drug efflux, increased repair of drug-induced DNA damage
Vancomycin, rifaximin
Microbial mechanisms of drug resistance
Drug inactivation.
Relationships between antibiotic use and emergence of drug-resistant microbes
< div class='tao-gold-member'>
![]()
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


Basic principles of antimicrobial therapy
Only gold members can continue reading. Log In or Register to continue
Get Clinical Tree app for offline access
