The GEnES Fingerprint—The Future
A framework to organize mental health data
For over a century, psychiatry has primarily utilized phenomenology—those occurrences and behaviors that are reported or are observed—as the basis for categorizing mental illness. Kraeplin organized the myriad presentations of mental conditions on the basis of what clinicians could observe and what patients described. Freud postulated mental forces and hypothetical constructs of mental functioning: the ego, id, and superego—entities that could not directly be observed. Psychoanalytic theory inferred their presence, but their loci could not be found. Observable symptoms of mental disorders were the result of interactions between these forces based on therapist
observation and patient verbal report. These two major thinkers shaped our conceptualizations and thinking during most of the 20th century. Even as we began to see many mental conditions as “brain illnesses” and began to categorize the symptom lists that make up the Diagnostic and Statistical Manual of Mental Disorders (DSM), we continue to rely on phenomenology in formulating our diagnostic categories.
observation and patient verbal report. These two major thinkers shaped our conceptualizations and thinking during most of the 20th century. Even as we began to see many mental conditions as “brain illnesses” and began to categorize the symptom lists that make up the Diagnostic and Statistical Manual of Mental Disorders (DSM), we continue to rely on phenomenology in formulating our diagnostic categories.
The age of tools
Three major advances—endophenotyping, neuroimaging, and genotyping—are changing our view of the brain, both literally and figuratively, and have begun to take center stage in the field of mental health. We now have the ability to observe neural structure and activity through an expanding series of brain-imaging techniques. We have also begun to genotype and categorize sequences of genetic material that occur with some predictability in both normal and abnormal mental states. Let us look at each of these issues more closely.
Endophenotypes
First used by Gottesman in 1991 and described in more detail by Gottesman and Gould in 2003 (1), the concept of endophenotypes makes a conceptual break with pure phenomenology by focusing on basic elemental concepts. Endophenotypes, which can be either biologic or psychological, are intermediate steps between genes and disease states. They are more precise than heterogeneous symptoms such as paranoia, hypomania, and mania, and more exact than symptom clusters or diagnoses such as bipolar I and II disorders or cyclothymia. Endophenotypes are specific, measurable biochemical elements such as the levels of serotonin in brain synapses or amygdala concentrations of catechol O-methyltransferase (COMT). They can also be measurable behavioral traits such as eye movement tracking, auditory startle response, and levels of risk avoidance. Endophenotypes, as more fundamental elements of brain functioning, are potentially components of, and/or signals for, diagnosable illnesses.
Critical endophenotypes are those processes related to a disorder but not necessarily symptoms of the disorder. They serve as proxy measures of the trait or disorder being studied, by connecting the external manifestation of the illness to underlying neural circuitry and genetic mechanisms. In order to satisfy criteria that are useful to clinical practice as well as research, critical endophenotypes must be reliable, stable, narrowly defined, and readily identifiable. Critical endophenotypes must also have phenotypic and genetic correlations, that is, they are specific to a particular disease (phenotypic correlation) and must be connected to underlying genetic mechanisms (genetic correlation). Lastly, these genetic and phenotypic correlations must relate to causality.
The concept of endophenotypes is immensely useful in virtually all areas of mental functioning, both normal and abnormal. As we focus on more basic elements of neural functioning, neurocircuitry, neurochemistry, and neurophysiology, there will be less direct focus on phenotypes and phenomenology.
Neuroimaging
Historically, the study of the physical brain was limited to viewing cadaver tissue. Newer processes of imaging the brain have given us in vivo methods to view brain structure and functioning. To use magnetic resonance imaging (MRI) as an example, we can noninvasively measure the size of neural tracts and brain centers in both normal and diseased individuals. We are able to statistically compare shapes and sizes of various brain loci between individuals, and in aggregate, between groups of individuals. It is possible to observe changes in brain structure and function over time, either in a quiescent state or when stimulated by biological and environmental perturbations. Functional magnetic resonance imaging (fMRI), single positron emission computerized tomography (SPECT), positron emission tomography (PET), blood oxygen-level-dependent magnetic resonance spectroscopy (BOLD), and other techniques provide us a glimpse of the brain as it operates normally, in disease, or in response to chemical and environmental stimuli. We are able to see what areas “light up” or “turn off” in response to tasks, stresses, and/or restriction of specific stimuli. By combining the images of multiple individuals, we can discern patterns of response and make hypotheses about neural functioning as it relates to disease. As we become more sophisticated in our imaging techniques, procedures such as diffusion tensor imaging (DTI), voxel-based morphometry (VBM), cortical surface-based analysis, and spherical harmonic (SPHARM) detection will explore specific circuits and fiber pathways more deeply and microscopically.
Genetic sequencing
Much work has occurred over the last decade in the area of human genomics. This painstaking $2.7 billion process culminated in 2003 in the accomplishment of the virtual, complete sequencing of the human genome. With the pattern of the genetic code established, processes such as positional cloning (linkage) and allelic association (linkage disequilibrium) as well as other techniques, are now unraveling the roles of specific genes. These roles include protein synthesis, gene regulation, and the downstream effects of gene products on other genes, neural circuits, and end organs in both normal and diseased individuals.
The need for a new system of data organization
Despite endophenotypic and genomic advances, the focus on phenomenology has persisted in our nosology. Although we are no longer constrained to
live behind the wall of phenomenology, we continue to categorize psychiatric data with outdated methodology. Although precise phenomenological nosology may be useful, it is hardly sufficient. No amount of phenomenological precision alone will directly provide precise information about the causality of mental illnesses. Even laboratory measurement of biochemical endophenotypes and neuroimaging are only likely to be intermediate steps in understanding the causes of mental disorders. Genetic testing of family inherited patterns and associated linkages as modified by exogenic and epigenetic phenomena, which are then correlated to biochemical endophenotypes and diagnostic syndromes, is a more rapid and accurate path to understanding the causes of mental disorders. We have spent decades obsessing about minor elements of phenotypic differences. We have clustered our symptoms into “diagnoses.” Although perhaps it has been the best that could have been accomplished at the time, this preoccupation is the slowest road to precise causality and symptomatic treatment. It may be less helpful to wrangle about whether a person has bipolar I or bipolar II disorder, or is cyclothymic than to separate and validate target symptoms, especially as we understand critical underlying endophenotypes and neurocircuitry that lead to these symptoms.
live behind the wall of phenomenology, we continue to categorize psychiatric data with outdated methodology. Although precise phenomenological nosology may be useful, it is hardly sufficient. No amount of phenomenological precision alone will directly provide precise information about the causality of mental illnesses. Even laboratory measurement of biochemical endophenotypes and neuroimaging are only likely to be intermediate steps in understanding the causes of mental disorders. Genetic testing of family inherited patterns and associated linkages as modified by exogenic and epigenetic phenomena, which are then correlated to biochemical endophenotypes and diagnostic syndromes, is a more rapid and accurate path to understanding the causes of mental disorders. We have spent decades obsessing about minor elements of phenotypic differences. We have clustered our symptoms into “diagnoses.” Although perhaps it has been the best that could have been accomplished at the time, this preoccupation is the slowest road to precise causality and symptomatic treatment. It may be less helpful to wrangle about whether a person has bipolar I or bipolar II disorder, or is cyclothymic than to separate and validate target symptoms, especially as we understand critical underlying endophenotypes and neurocircuitry that lead to these symptoms.
Any new classification and model of mental illness will need to conform to the following:
Be based on data beyond pure phenomenology.
Link biopsychosocial information to genetic and neurophysiologic data.
Connect new research data to currently used diagnostic concepts.
Facilitate the redefinition and restructuring of current diagnostic concepts and symptoms into more basic elements.
Identify consistent data categories that can be organized into a standardized searchable database to identify connections between disparate forms of data regarding a particular disease entity or patient.
A new conceptualization of the continuum from genetics to diagnosis—the GEnES fingerprint
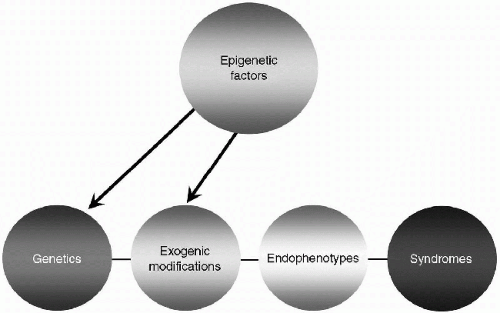
This chapter proposes a new framework for classifying psychiatric data that includes all current information sources in a simple, yet flexible model of mental processes called the GEnES Fingerprint. Beyond creating a system for organizing and collecting data on psychiatric illnesses, this model also easily adapts itself to providing a biologic and psychological understanding of variations of personality traits, both “abnormal” and “normal.” It is a model that allows for the input of the effects of nongenetic sources such as environment, parenting, culture, medication, and nonmedication treatments such as psychotherapy and electroconvulsive therapy (ECT).
By spelling “GEnES” this way, the capital letters suggest the four primary types of data in a continuum—Genetics, Exogenic modifications, Endophenotypes, and Syndromes. Each data set is separate, and connects to the others in
a linear manner shown in simplified form in Figure 2.1. As the model is elucidated, the GEnES Fingerprint of each psychiatric condition will ultimately be found to be as unique as a person’s fingerprint. This level of precision requires sophisticated measurement tools. Some disease fingerprints will have more specificity than others. It will take time to develop each data set and initially there may be a complete absence of information or only hypothetical links from one data set to the next.
a linear manner shown in simplified form in Figure 2.1. As the model is elucidated, the GEnES Fingerprint of each psychiatric condition will ultimately be found to be as unique as a person’s fingerprint. This level of precision requires sophisticated measurement tools. Some disease fingerprints will have more specificity than others. It will take time to develop each data set and initially there may be a complete absence of information or only hypothetical links from one data set to the next.
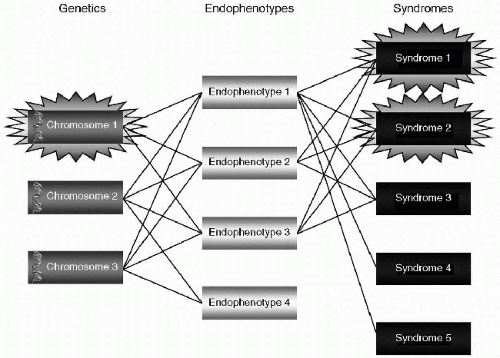
Further refining of the four primary data types includes the development of distinct subgroups within each type of data—Genes, Exogenic modifications, Endophenotypes, and Syndromes. These subgroups are labeled with consistent numbering and abbreviations (see Tables 2.1, 2.2). By labeling data with its category and subset number, a coherent stable system is created for researchers, clinicians, and educators to collect, organize, analyze, and teach mental health data. As information is ultimately connected to elements within the other data sets, underlying causal mechanisms will be connected to the
phenotypic expressions that we call personality traits, personality disorders, diagnoses, and mental illnesses. Although many elements of the fingerprint may not yet be known, the methodology will more clearly direct our thinking toward formulating more precise research, as well as more specific treatments and therapies.
phenotypic expressions that we call personality traits, personality disorders, diagnoses, and mental illnesses. Although many elements of the fingerprint may not yet be known, the methodology will more clearly direct our thinking toward formulating more precise research, as well as more specific treatments and therapies.
TABLE 2.1 Genetic data sets | ||||||
|---|---|---|---|---|---|---|
|
TABLE 2.2 Endophenotypic data sets | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||
Discussion of the model components
Type 1 data—genetics
In the article Psychiatric Genetics: a Methodological Critique, Kendler describes four paradigms of the role of genetic factors in the etiology of psychiatric disorders (2). He describes both basic and advanced genetic epidemiology, which are statistical analyses of an individual’s risk or liability of psychiatric illness. These risks result from familial hereditary effects as measured in twin or adoption studies. Other researchers refer to this form of genetic study as positional cloning (linkage) in which chromosomal abnormalities located in genome-wide scans are “linked” to a disorder. In this methodology, there is a search for relationships between loci of deoxyribonucleic acid (DNA) nucleotide markers and diseases, as they present in high-risk families. Results yield large chromosomal regions of multiple nucleotide pairs that appear linked to a particular condition, thereby yielding data that is often relatively nonspecific. When further study on these lengthy DNA sequences is congruent (3,4) the genomic region for a particular disease such as bipolar disorder can be narrowed down to a smaller sequence or possibly an isolated nucleotide pair (see Figure 2.2).
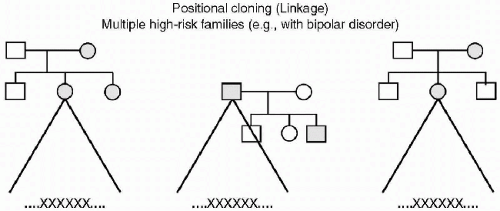 Figure 2.2 XXXXXX—An abnormal sequence of DNA (which may be quite large) that appears in multiple families. |
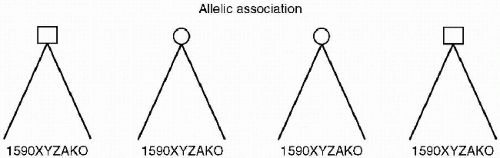
Kendler goes on to describe gene identification research that seeks to find more precise areas of genetic abnormality that can then be tied to neuroanatomic or neurophysiologic changes, and ultimately to phenotypes and psychiatric illness. Other researchers use the term Allelic Association or Linkage Disequilibrium to describe this type of genetic research. In this methodology, there is a search for relationships between specific changes in a genetic sequence (alleles) that code for the formation of neuroanatomic structures and specific neurophysiologic processes. It is hoped that these expressions of genetic material can be tied to disease phenotypes in general and/or specific psychiatric disorders in unrelated individuals. This data can be potentially exact, but in order to be clinically useful, the data needs to be correlated to endophenotypes and symptoms through the identification of neural circuits related to the disorder (see Figure 2.3).
Each of these paradigms has strengths and weaknesses, and results in different forms of data. They are shown in Table 2.1 in the subsequent text.
Genetic 1 and 2 data are useful in understanding the ultimate causality of psychiatric illnesses, neurophysiologic deficits, and strengths. Other methodologies to identify genetic contributions to mental illnesses may be elucidated, and as genetic investigation progresses, further categories will likely be created.
It is tempting to assume that once the genetic coding is elucidated using the methodologies described in the preceding text we will have a clear biologic explanation of bipolar disorder and other psychiatric illnesses. This assumes a rather simplistic genetic model that Kendler has termed the essentialist gene model (5). As was put forth in the model of Mendelian genetics, this postulates a very strong connection between disruption of gene function and the ultimate illness. There are, however, many other factors beyond simple genetic coding that may influence the ultimate syndromal outcome of genetic sequences. Although the role of these processes is only rudimentarily understood, the model can account for them.
Gene products are not always consistent—exogenic modifications
As we now know, a variety of influences may impact the ultimate output of genetic DNA. These can include regulation of the quantity of protein made by a gene, modification of the genetic transcription process, and effects of messenger ribonucleic acid (RNA) to name a few. These processes, in different ways, alter the proteins created from the underlying genetic sequencing. If these processes emerge as significant links in the chain from DNA to endophenotypes and syndromes (and it is probable that they do), simply identifying abnormal genetic sequencing may only be part of the causal chain to psychiatric illnesses.
The elements of gene regulation and expression, as well as elements of design, exist “downstream” of the genetic coding itself; that is, beyond DNA. We now know that the “one gene equals one enzyme” hypothesis is untrue. In the human genome, 75% of human genes are alternatively spliced, leading to alternative forms of each gene (6). Because of this, the same gene (i.e., the same nucleotide sequence) can produce different messenger RNA transcripts that can result in different proteins (7).
Gene sequences at a distance and separate from an individual gene locus can regulate the amount of protein produced and the actual nature of the protein produced by that locus. Because of multiple promoters and regulators of gene activity, different protein variants can emerge from a single gene at different times in different tissues. Some of these alterations occur because of different splicing of DNA sequences and others occur because of RNA editing. In the latter activity, the posttranscriptional RNA sequence is edited from that originally encoded by the DNA and can, in some cases, alter the structure of the expressed protein (8).
The impact of RNA in relation to the ultimate expression of proteins adds another layer of complexity to the overly simplistic “one gene-one
protein” theory. There are at least two forms of RNA that are noncoding. The first is housekeeping RNA involved in splicing and translation. The second is regulatory RNA that alters gene expression through transcriptional and posttranscriptional mechanisms. In at least one species of plants (9), elements of design reside outside DNA in ancestral RNA. If such elements were found to be present in humans, an entirely new level of information would emerge, which could be further factored into the ultimate explanation and understanding of mental health phenotypes, and would introduce novel sites for mental health treatments.
protein” theory. There are at least two forms of RNA that are noncoding. The first is housekeeping RNA involved in splicing and translation. The second is regulatory RNA that alters gene expression through transcriptional and posttranscriptional mechanisms. In at least one species of plants (9), elements of design reside outside DNA in ancestral RNA. If such elements were found to be present in humans, an entirely new level of information would emerge, which could be further factored into the ultimate explanation and understanding of mental health phenotypes, and would introduce novel sites for mental health treatments.
In the GEnES Fingerprint, these chemical processes that occur “downstream” from DNA sequencing and transcription are combined under the rubric of “exogenic modifications.” We are just beginning to understand the role of these various processes in the development of endophenotypes and ultimately in mental health syndromes, but the pictorial model shown in Figures 2.4 and 2.6 highlight the likelihood that a strict gene-to-endophenotype path is unlikely. Although it remains purely speculative at this time in our understanding, one can postulate that the epigenetic factors shown in this diagram exert their influence at least in part through these exogenic mechanisms that modify the protein output from DNA. In this way, the epigenetic factors influence the emergence of endophenotypes and ultimately syndromes.
Scientific advancement is dependent on expanding the current purely phenomenologic model to one that incorporates the other factors. There
is little doubt that as we collect, organize, compare, and connect various elements of biologic and psychological data, refinements of the GEnES model or other novel models will emerge. The flexibility of the GEnES Fingerprint model is that it allows for input and refinement that can easily be incorporated as new information is uncovered.
is little doubt that as we collect, organize, compare, and connect various elements of biologic and psychological data, refinements of the GEnES model or other novel models will emerge. The flexibility of the GEnES Fingerprint model is that it allows for input and refinement that can easily be incorporated as new information is uncovered.
Endophenotypes
As defined in the preceding text, endophenotypes are intermediate, more basic steps between genes and diseases. This type of data is further broken down into subtypes as shown in Table 2.2 and discussed in the subsequent text. This section presents examples of potential endophenotypes with some current research validity; however, no attempt is made to suggest that these are, in fact, criteria which will ultimately have clinical, heuristic, or therapeutic usefulness. As discussed later, there are relatively few endophenotypes that specifically relate to elevated mood. Therefore, illustrative examples are presented from other various mental conditions and research.
It is predictable that data will fit several endophenotypes and will be catalogued in more than one location within the model. One would expect that as data sources are entered, recorded, and connected, further refinements and subtyping of endophenotypes will occur.
Biochemical endophenotypes
Examples include COMT activity in patients with rapid cycling bipolar disorder (10) or basal and stimulated cortical gamma-aminobutyric acid (GABA) and calcium levels in olfactory receptor neurons of individuals with bipolar disorder (11). Here, the levels of serotonin resulting from a mutation in the gene for human tryptophan hydroxylase-2 (12) and the pituitary response of adrenocorticotropic hormone (ACTH) in the brain to dexamethasone are also catalogued (13).
Infectious endophenotypes
Endocrinologic endophenotypes
This subcategory includes all data regarding the effects of endocrine systems in the body such as the thyroid, adrenal glands, and other hormone-secreting systems. The effects of insulin deficiency on Alzheimer’s disease (17), the psychological effects of excess glucocorticoids on psychological states in Cushing’s syndrome (18), and research on psychosexual development and androgen insensitivity syndrome (19) can also be categorized here.
Neurobehavioral endophenotypes
Inflammatory endophenotypes
Neuroanatomic endophenotypes
Data here includes gross measurement of brain structures such as hippocampal and superior temporal gyrus volumes, as well as measurement of dopamine and serotonin transporter densities. Also included are the effects of neurologic and non-neurologic illnesses (e.g., cancer, multiple sclerosis, blunt brain trauma, and others). Also listed are the neuroanatomic correlates of the psychopathologic components of major depressive disorder (24), the amount, distribution and ratio of complexin I and II in the hippocampi of schizophrenics (25), and the balance between excitatory parietal cell firing and GABA-mediated inhibition (26).
Neuropsychological endophenotypes
Examples here include assessment of P50 suppression and prepulse inhibition of startle response (27) and measurements of emotional processing such as performance on the Affective Go/No Go test in the face of chronic methylenedioxymethamphetamine (MDMA “ecstasy”) use (28). This subgroup also includes configured self-reports such as reports of subjective experiences (29), the effect of personality traits on other endophenotypes or behaviors such as suicidal actions (30,31) and using a self-report depression scale to identify remission (32). Data on neuropsychological impairments in tasks measuring episodic memory, verbal fluency, psychomotor speed, and executive functioning in anxious patients (33) would be listed here as well as under the cognitive subtype.
Cognitive endophenotypes
Data here includes tests of visual executive processing and selective attention indicated by continuous performance tasks such as the forced choice visual oddball task (34). Additionally, cognitive testing results of individuals at high risk for schizophrenia or Alzheimer’s disease, such as assessments of working memory, cognitive dysmetria, and the fMRI of persons performing working
memory tasks (35) are categorized here. Information about the latter can include scores on the Boston Naming Test and the Block Design subtest of the Wechsler Intelligence Scale for children-Revised (36).
memory tasks (35) are categorized here. Information about the latter can include scores on the Boston Naming Test and the Block Design subtest of the Wechsler Intelligence Scale for children-Revised (36).
Immunologic endophenotypes
Here, information about the neural response to infectious agents, vaccinations, and autoimmune responses of the body are listed. Examples include data related to the PANDAS (psychiatric autoimmune neuropsychiatric disorders associated with streptococcal infections) and their possible connection to obsessive-compulsive disorder (OCD) and tic disorders such as Tourette’s syndrome (37). Identifying GABAergic synapses in the brain using antisera to GABA (38), the possibility of “vaccinating” individuals against a neurodegenerative disorder such as Alzheimer’s disease (39) and immunoglobulin treatment strategies for autism (40,41,42) are also categorized here.
Other endophenotypes
Before moving on to the concepts of syndromes as defined in this model, it must be mentioned that there are considerable psychiatric data that fall under the rubric of measurement of endophenotypes. These data are not endophenotypes per se but are data generated by tools that document the presence of, and quantify the extent of endophenotypes in humans and animals. There are five such data types, as shown in Table 2.3.
Imaging
These data include a variety of neuroimaging results from currently used and potentially new neuroimaging technology, such as SPECT, PET, fMRI, BOLD, VBM scans, and others. There can be scans of individuals, composite data from multiple scans of a group of individuals, or comparative scans of the same individual at different times and under differing conditions.
TABLE 2.3 Measurements of endophenotypes | |||||
|---|---|---|---|---|---|
|
Specific examples are fMRI data related to smooth pursuit eye movement tasks in schizophrenia when nicotine is introduced (43), MRI hyperintensities in individuals with depression (44), PET scans during presleep wakefulness and non-REM (rapid eye movement) sleep (45), SPECT scans of dopamine transporter densities in patients with OCD (46), and voxel-based morphometry of gray matter in patients with bipolar disorder (47).
Electrophysiologic measurements
These data include baseline or sleep-deprived electroencephalogram (EEG) tracings, evoked potential, and other measurements of brain electrical activity in the resting or stimulated state. Examples could also include eye-tracking dysfunction and deficits in P50 event-related potential inhibition in the auditory-click-conditioning test paradigm and saccadic inhibition deficits (48) as well as measurement of auditory startle response through eye blink electromyograms and skin conductance response (49).
Effects of pharmacologic interventions
This subcategory contains information relating to the effects of specific pharmacologic agents on brain functioning, neurophysiologic or neuroanatomic substrates. It can be organized by both specific medications and groupings of medications (if the latter are shown to provide consistently similar neural effects). For example, impaired GABA neural response to acute benzodiazepine administration (50), the occurrence of tachyphylaxis (loss of effect) during antidepressant treatment (51) and the safety profile of various medications in the pregnant patient are categorized in this subtype. Also codified here would be the rates of response of anxiety and depression measures in patients to paroxetine (52) or the prediction of drug response based on genetic polymorphisms (53). This latter procedure connects pharmacologic response rates to genetic haplotypes.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


Get Clinical Tree app for offline access